Additional material for the article “Coverage-based
resampling: Building robust consolidated decision trees”
This page
contains the additional material related to the work presented in "Igor
Ibarguren, Jesús M. Pérez, Javier Muguerza, Ibai Gurrutxaga and Olatz
Arbelaitz, Coverage-based resampling: Building robust consolidated decision
trees, Knowledge-Based Systems, Vol. 79, May 2015, pp. 51-67". It is
available online at http://dx.doi.org/10.1016/j.knosys.2014.12.023.
Index
1 Data set characteristics
2 Subsample numbers by data set
to achieve the selected coverage values
3 Results of the Wilcoxon Signed
Ranks test regarding comparisons between subsample sizes
4 Execution times for CTC
5 Average Results achieved by CTC
6 Comparison between CTC and
methods to tackle class imbalance
1
Data set characteristics
The tables in this section summarize the characteristics of each data
set used in the article. Table 1 refers to standard data sets while Table 2 refers to imbalanced data sets.
|
Data set |
#Atts. |
#Examples |
#Classes |
%min |
%maj |
Size of
Min. Class |
Size of
Maj. Class |
|
lymphography |
18 |
148 |
4 |
1.36% |
54.73% |
2 |
81 |
|
ecoli |
7 |
336 |
8 |
0.6% |
42.56% |
2 |
143 |
|
car |
6 |
1728 |
4 |
3.77% |
70.03% |
65 |
1210 |
|
nursery |
8 |
1296 |
5 |
0.08% |
33.34% |
1 |
432 |
|
cleveland |
13 |
297 |
5 |
4.38% |
53.88% |
13 |
160 |
|
zoo |
17 |
101 |
7 |
3.97% |
40.6% |
4 |
41 |
|
glass |
9 |
214 |
6 |
4.21% |
35.52% |
9 |
76 |
|
flare |
10 |
1066 |
6 |
4.04% |
31.06% |
43 |
331 |
|
abalone |
8 |
418 |
22 |
0.24% |
16.51% |
1 |
69 |
|
balance |
4 |
625 |
3 |
7.84% |
46.08% |
49 |
288 |
|
dermatology |
33 |
358 |
6 |
5.59% |
31.01% |
20 |
111 |
|
hepatitis |
19 |
80 |
2 |
16.25% |
83.75% |
13 |
67 |
|
newthyroid |
5 |
215 |
3 |
13.96% |
69.77% |
30 |
150 |
|
haberman |
3 |
306 |
2 |
26.48% |
73.53% |
81 |
225 |
|
breast |
9 |
277 |
2 |
29.25% |
70.76% |
81 |
196 |
|
german |
20 |
1000 |
2 |
30% |
70% |
300 |
700 |
|
wisconsin |
9 |
630 |
2 |
34.61% |
65.4% |
218 |
412 |
|
contraceptive |
9 |
1473 |
3 |
22.61% |
42.71% |
333 |
629 |
|
tictactoe |
9 |
958 |
2 |
34.66% |
65.35% |
332 |
626 |
|
pima |
8 |
768 |
2 |
34.9% |
65.11% |
268 |
500 |
|
magic |
10 |
1902 |
2 |
35.13% |
64.88% |
668 |
1234 |
|
wine |
13 |
178 |
3 |
26.97% |
39.89% |
48 |
71 |
|
bupa |
6 |
345 |
2 |
42.03% |
57.98% |
145 |
200 |
|
heart |
13 |
270 |
2 |
44.45% |
55.56% |
120 |
150 |
|
australian |
14 |
690 |
2 |
44.5% |
55.51% |
307 |
383 |
|
crx |
15 |
653 |
2 |
45.33% |
54.68% |
296 |
357 |
|
vehicle |
18 |
846 |
4 |
23.53% |
25.77% |
199 |
218 |
|
penbased |
16 |
1100 |
10 |
9.55% |
10.46% |
105 |
115 |
|
ring |
20 |
740 |
2 |
49.6% |
50.41% |
367 |
373 |
|
iris |
4 |
150 |
3 |
33.34% |
33.34% |
50 |
50 |
|
Mean |
11.77 |
638.93 |
4.27 |
21% |
50% |
139 |
319.93 |
|
StdDev |
6.44 |
493.55 |
3.9 |
16.41% |
18.09% |
158.42 |
306.8 |
|
Median |
9.5 |
521.5 |
3 |
23% |
54% |
73 |
209 |
Table 1: Description of standard data sets.
|
Data set |
#Atts. |
#Examples |
Imbalance |
Size of
Min. Class |
Size of Maj.
Class |
|
Abalone19 |
8 |
4174 |
0.77% |
32 |
4142 |
|
Yeast6 |
8 |
1484 |
2.49% |
37 |
1447 |
|
Yeast5 |
8 |
1484 |
2.96% |
44 |
1440 |
|
Yeast4 |
8 |
1484 |
3.43% |
51 |
1433 |
|
Yeast2vs8 |
8 |
482 |
4.15% |
20 |
462 |
|
Glass5 |
9 |
214 |
4.2% |
9 |
205 |
|
Abalone9vs18 |
8 |
731 |
5.65% |
41 |
690 |
|
Glass4 |
9 |
214 |
6.07% |
13 |
201 |
|
Ecoli4 |
7 |
336 |
6.74% |
23 |
313 |
|
Glass2 |
9 |
214 |
8.78% |
19 |
195 |
|
Vowel0 |
13 |
988 |
9.01% |
89 |
899 |
|
Page-blocks0 |
10 |
5472 |
10.23% |
560 |
4912 |
|
Ecoli3 |
7 |
336 |
10.88% |
37 |
299 |
|
Yeast3 |
8 |
1484 |
10.98% |
163 |
1321 |
|
Glass6 |
9 |
214 |
13.55% |
29 |
185 |
|
Segment0 |
19 |
2308 |
14.26% |
329 |
1979 |
|
Ecoli2 |
7 |
336 |
15.48% |
52 |
284 |
|
New-thyroid1 |
5 |
215 |
16.28% |
35 |
180 |
|
New-thyroid2 |
5 |
215 |
16.89% |
36 |
179 |
|
Ecoli1 |
7 |
336 |
22.92% |
77 |
259 |
|
Vehicle0 |
18 |
846 |
23.64% |
200 |
646 |
|
Glass0123vs456 |
9 |
214 |
23.83% |
51 |
163 |
|
Haberman |
3 |
306 |
27.42% |
84 |
222 |
|
Vehicle1 |
18 |
846 |
28.37% |
240 |
606 |
|
Vehicle2 |
18 |
846 |
28.37% |
240 |
606 |
|
Vehicle3 |
18 |
846 |
28.37% |
240 |
606 |
|
Yeast1 |
8 |
1484 |
28.91% |
429 |
1055 |
|
Glass0 |
9 |
214 |
32.71% |
70 |
144 |
|
Iris0 |
4 |
150 |
33.33% |
50 |
100 |
|
Pima |
8 |
768 |
34.84% |
268 |
500 |
|
Ecoli0vs1 |
7 |
220 |
35% |
77 |
143 |
|
Wisconsin |
9 |
683 |
35% |
239 |
444 |
|
Glass1 |
9 |
214 |
35.51% |
76 |
138 |
|
Mean |
9.39 |
919.94 |
17.61% |
120 |
799.94 |
|
StdDev |
4.17 |
1151.99 |
11.70% |
132.19 |
1800 |
|
Median |
8 |
482 |
15.48% |
52 |
444 |
Table 2: Description of imbalanced data sets.
2
Subsample numbers by data set to achieve the selected coverage values
The tables in this section show the number of subsamples computed for
each data set for any of the used coverage values. Table 3 and Table 4 refer to standard data sets, sizeOfMinClass
and maxSize subsamples respectively. Table 5 and Table 6 refer to imbalanced data sets, sizeOfMinClass
and maxSize subsamples respectively.
For standard datasets the MinCover column
represent the minimum number of examples of each class as stated by the rule
and exceptions in the methodology section of the article. The data sets where
the size of classes in the subsamples is enforced by the MinCover as opposed to
the size of the minority class are stressed in bold.
For imbalanced data sets preprocessed with
SMOTE, only the total example number and the size of the minority class change
from the data sets without the preprocessing. In these data sets the minority
class has been oversampled with SMOTE until it has the same size as the
majority class.
|
|
Original |
Training
sample |
Subsample |
|
Coverage
sizeOfMinClass |
|||||||||||||||
|
Data set |
Size |
#Class |
%Min |
Size |
Min.
Class Size |
MinCover[1] |
Maj.
Class Size |
Size |
|
N_S |
||||||||||
|
|
|
|
|
|
NS=3 |
10% |
20% |
30% |
40% |
50% |
75% |
90% |
95% |
99% |
99.9% |
|||||
|
lymphography |
148 |
4 |
1.36% |
119 |
2 |
2 |
66 |
8 |
3.04% |
3 |
4 |
8 |
12 |
17 |
23 |
46 |
75 |
98 |
150 |
225 |
|
ecoli |
336 |
8 |
0.6% |
269 |
2 |
3 |
115 |
24 |
2.61% |
3 |
4 |
9 |
14 |
20 |
27 |
53 |
88 |
114 |
175 |
262 |
|
car |
1728 |
4 |
3.77% |
1383 |
53 |
14 |
969 |
108 |
2.79% |
3 |
4 |
8 |
13 |
19 |
25 |
50 |
82 |
107 |
163 |
245 |
|
nursery |
1296 |
5 |
0.08% |
1037 |
1 |
11 |
346 |
55 |
3 |
4 |
7 |
12 |
16 |
22 |
43 |
72 |
93 |
143 |
214 |
|
|
cleveland |
297 |
5 |
4.38% |
238 |
11 |
3 |
129 |
30 |
4.66% |
3 |
3 |
5 |
8 |
11 |
15 |
30 |
49 |
63 |
97 |
146 |
|
zoo |
101 |
7 |
3.97% |
81 |
4 |
1 |
33 |
14 |
6.07% |
3 |
3 |
4 |
6 |
9 |
12 |
23 |
37 |
48 |
74 |
111 |
|
glass |
214 |
6 |
4.21% |
172 |
8 |
2 |
62 |
24 |
6.46% |
3 |
3 |
4 |
6 |
8 |
11 |
21 |
35 |
45 |
70 |
104 |
|
flare |
1066 |
6 |
4.04% |
853 |
35 |
9 |
265 |
108 |
6.8% |
3 |
3 |
4 |
6 |
8 |
10 |
20 |
33 |
43 |
66 |
99 |
|
abalone |
418 |
22 |
0.24% |
335 |
1 |
4 |
56 |
88 |
7.15% |
3 |
3 |
4 |
5 |
7 |
10 |
19 |
32 |
41 |
63 |
94 |
|
balance |
625 |
3 |
7.84% |
500 |
40 |
5 |
231 |
60 |
8.66% |
3 |
3 |
3 |
4 |
6 |
8 |
16 |
26 |
34 |
51 |
77 |
|
dermatology |
358 |
6 |
5.59% |
287 |
17 |
3 |
89 |
54 |
10.12% |
3 |
3 |
3 |
4 |
5 |
7 |
14 |
22 |
29 |
44 |
65 |
|
hepatitis |
80 |
2 |
16.25% |
64 |
11 |
1 |
54 |
12 |
11.12% |
3 |
3 |
3 |
4 |
5 |
6 |
12 |
20 |
26 |
40 |
59 |
|
newthyroid |
215 |
3 |
13.96% |
172 |
24 |
2 |
120 |
36 |
10% |
3 |
3 |
3 |
4 |
5 |
7 |
14 |
22 |
29 |
44 |
66 |
|
haberman |
306 |
2 |
26.48% |
245 |
65 |
3 |
181 |
66 |
18.24% |
3 |
3 |
3 |
3 |
3 |
4 |
7 |
12 |
15 |
23 |
35 |
|
breast |
277 |
2 |
29.25% |
222 |
65 |
3 |
158 |
66 |
20.89% |
3 |
3 |
3 |
3 |
3 |
3 |
6 |
10 |
13 |
20 |
30 |
|
german |
1000 |
2 |
30% |
800 |
240 |
8 |
560 |
240 |
21.43% |
3 |
3 |
3 |
3 |
3 |
3 |
6 |
10 |
13 |
20 |
29 |
|
wisconsin |
630 |
2 |
34.61% |
504 |
175 |
6 |
330 |
176 |
26.67% |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
8 |
10 |
15 |
23 |
|
contraceptive |
1473 |
3 |
22.61% |
1179 |
267 |
12 |
504 |
402 |
26.59% |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
8 |
10 |
15 |
23 |
|
tictactoe |
958 |
2 |
34.66% |
767 |
266 |
8 |
502 |
266 |
26.5% |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
8 |
10 |
15 |
23 |
|
pima |
768 |
2 |
34.9% |
615 |
215 |
7 |
401 |
216 |
26.94% |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
8 |
10 |
15 |
23 |
|
magic |
1902 |
2 |
35.13% |
1522 |
535 |
16 |
988 |
536 |
27.13% |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
8 |
10 |
15 |
22 |
|
wine |
178 |
3 |
26.97% |
143 |
39 |
2 |
58 |
60 |
34.49% |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
6 |
8 |
11 |
17 |
|
bupa |
345 |
2 |
42.03% |
276 |
116 |
3 |
160 |
116 |
36.25% |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
6 |
7 |
11 |
16 |
|
heart |
270 |
2 |
44.45% |
216 |
96 |
3 |
120 |
96 |
40% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
6 |
10 |
14 |
|
australian |
690 |
2 |
44.5% |
552 |
246 |
6 |
307 |
246 |
40.07% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
6 |
9 |
14 |
|
crx |
653 |
2 |
45.33% |
523 |
238 |
6 |
286 |
238 |
41.61% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
6 |
9 |
13 |
|
vehicle |
846 |
4 |
23.53% |
677 |
160 |
7 |
175 |
320 |
45.72% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
5 |
8 |
12 |
|
penbased |
1100 |
10 |
9.55% |
880 |
84 |
9 |
92 |
420 |
45.66% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
5 |
8 |
12 |
|
ring |
740 |
2 |
49.6% |
592 |
294 |
6 |
299 |
294 |
49.17% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
5 |
7 |
11 |
|
iris |
150 |
3 |
33.34% |
120 |
40 |
2 |
40 |
60 |
50% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
5 |
7 |
10 |
|
Mean |
638.94 |
4.27 |
22% |
511.44 |
111.67 |
5.57 |
256.54 |
147.97 |
22% |
3 |
4 |
4 |
5 |
7 |
8 |
15 |
24 |
31 |
47 |
70 |
|
StdDev |
493.55 |
3.96 |
16% |
394.87 |
126.76 |
3.92 |
245.53 |
139.38 |
53% |
0.18 |
0.77 |
2.22 |
3.71 |
5.68 |
9.46 |
17.84 |
26.94 |
37.25 |
56.61 |
72.13 |
|
Median |
521.5 |
3 |
23.07% |
417.5 |
59 |
4.5 |
167.5 |
92 |
21.16% |
3 |
3 |
3 |
3 |
3 |
3 |
6 |
10 |
13 |
20 |
29.5 |
Table 3: sizeOfMinClass sized
subsample numbers for standard data sets.
|
|
Original |
Training
sample |
Subsample |
|
Coverage
maxSize |
|||||||||||||||
|
Data set |
Size |
#Class |
%Min |
Size |
Min.
Class Size |
MinCover[2] |
Maj.
Class Size |
Size |
|
N_S |
||||||||||
|
|
|
|
|
|
N_S=3 |
10% |
20% |
30% |
40% |
50% |
75% |
90% |
95% |
99% |
99.9% |
|||||
|
lymphography |
148 |
4 |
1.36% |
119 |
2 |
2 |
66 |
12 |
4.55% |
3 |
3 |
5 |
8 |
11 |
15 |
30 |
50 |
65 |
99 |
149 |
|
ecoli |
336 |
8 |
0.6% |
269 |
2 |
3 |
115 |
48 |
5.22% |
3 |
3 |
5 |
7 |
10 |
13 |
26 |
43 |
56 |
86 |
129 |
|
car |
1728 |
4 |
3.77% |
1383 |
53 |
14 |
969 |
212 |
5.47% |
3 |
3 |
4 |
7 |
10 |
13 |
25 |
41 |
54 |
82 |
123 |
|
nursery |
1296 |
5 |
0.08% |
1037 |
1 |
11 |
346 |
105 |
6.07% |
3 |
3 |
4 |
6 |
9 |
12 |
23 |
37 |
48 |
74 |
111 |
|
cleveland |
297 |
5 |
4.38% |
238 |
11 |
3 |
129 |
55 |
8.53% |
3 |
3 |
3 |
5 |
6 |
8 |
16 |
26 |
34 |
52 |
78 |
|
zoo |
101 |
7 |
3.97% |
81 |
4 |
1 |
33 |
28 |
12.13% |
3 |
3 |
3 |
3 |
4 |
6 |
11 |
18 |
24 |
36 |
54 |
|
glass |
214 |
6 |
4.21% |
172 |
8 |
2 |
62 |
48 |
12.91% |
3 |
3 |
3 |
3 |
4 |
6 |
11 |
17 |
22 |
34 |
51 |
|
flare |
1066 |
6 |
4.04% |
853 |
35 |
9 |
265 |
210 |
13.21% |
3 |
3 |
3 |
3 |
4 |
5 |
10 |
17 |
22 |
33 |
49 |
|
abalone |
418 |
22 |
0.24% |
335 |
1 |
4 |
56 |
154 |
12.5% |
3 |
3 |
3 |
3 |
4 |
6 |
11 |
18 |
23 |
35 |
52 |
|
balance |
625 |
3 |
7.84% |
500 |
40 |
5 |
231 |
120 |
17.32% |
3 |
3 |
3 |
3 |
3 |
4 |
8 |
13 |
16 |
25 |
37 |
|
dermatology |
358 |
6 |
5.59% |
287 |
17 |
3 |
89 |
102 |
19.11% |
3 |
3 |
3 |
3 |
3 |
4 |
7 |
11 |
15 |
22 |
33 |
|
hepatitis |
80 |
2 |
16.25% |
64 |
11 |
1 |
54 |
22 |
20.38% |
3 |
3 |
3 |
3 |
3 |
4 |
7 |
11 |
14 |
21 |
31 |
|
newthyroid |
215 |
3 |
13.96% |
172 |
24 |
2 |
120 |
72 |
20% |
3 |
3 |
3 |
3 |
3 |
4 |
7 |
11 |
14 |
21 |
31 |
|
haberman |
306 |
2 |
26.48% |
245 |
65 |
3 |
181 |
130 |
35.92% |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
6 |
7 |
11 |
16 |
|
breast |
277 |
2 |
29.25% |
222 |
65 |
3 |
158 |
130 |
41.14% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
6 |
9 |
14 |
|
german |
1000 |
2 |
30% |
800 |
240 |
8 |
560 |
480 |
42.86% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
6 |
9 |
13 |
|
wisconsin |
630 |
2 |
34.61% |
504 |
175 |
6 |
330 |
350 |
53.04% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
4 |
7 |
10 |
|
contraceptive |
1473 |
3 |
22.61% |
1179 |
267 |
12 |
504 |
801 |
52.98% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
4 |
7 |
10 |
|
tictactoe |
958 |
2 |
34.66% |
767 |
266 |
8 |
502 |
532 |
52.99% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
4 |
7 |
10 |
|
pima |
768 |
2 |
34.9% |
615 |
215 |
7 |
401 |
430 |
53.62% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
6 |
9 |
|
magic |
1902 |
2 |
35.13% |
1522 |
535 |
16 |
988 |
1070 |
54.15% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
6 |
9 |
|
wine |
178 |
3 |
26.97% |
143 |
39 |
2 |
58 |
117 |
67.25% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
7 |
|
bupa |
345 |
2 |
42.03% |
276 |
116 |
3 |
160 |
232 |
72.5% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
6 |
|
heart |
270 |
2 |
44.45% |
216 |
96 |
3 |
120 |
192 |
80% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
|
australian |
690 |
2 |
44.5% |
552 |
246 |
6 |
307 |
492 |
80.14% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
|
crx |
653 |
2 |
45.33% |
523 |
238 |
6 |
286 |
476 |
83.22% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
|
vehicle |
846 |
4 |
23.53% |
677 |
160 |
7 |
175 |
640 |
91.43% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
|
penbased |
1100 |
10 |
9.55% |
880 |
84 |
9 |
92 |
840 |
91.31% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
|
ring |
740 |
2 |
49.6% |
592 |
294 |
6 |
299 |
588 |
98.33% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
|
iris[3] |
150 |
3 |
33.34% |
120 |
40 |
2 |
40 |
66 |
55% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
6 |
9 |
|
Mean |
638.94 |
4.27 |
22% |
511.44 |
111.67 |
5.57 |
256.54 |
291.8 |
43% |
3 |
3 |
4 |
4 |
5 |
6 |
9 |
13 |
16 |
24 |
36 |
|
StdDev |
493.55 |
3.96 |
16% |
394.87 |
126.76 |
3.92 |
245.53 |
280.88 |
31% |
0 |
0 |
0.55 |
1.43 |
2.42 |
3.53 |
7.96 |
13.65 |
18.08 |
27.82 |
41.93 |
|
Median |
521.5 |
3 |
23.07% |
417.5 |
59 |
4.5 |
167.5 |
173 |
42% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
6 |
9 |
13.5 |
Table 4: maxSize sized subsample numbers for
standard data sets.
|
|
Original |
Training sample |
Subsample |
|
Coverage sizeOfMinClass |
|||||||||||||
|
Data set |
Size |
%Min |
Size |
Min. Class Size |
Maj. Class Size |
Size |
|
N_S |
||||||||||
|
|
|
|
|
|
N_S=3 |
10% |
20% |
30% |
40% |
50% |
75% |
90% |
95% |
99% |
99.9% |
|||
|
Abalone19 |
4174 |
0.77 |
3340 |
26 |
3314 |
26 |
0.39% |
3 |
27 |
57 |
91 |
130 |
177 |
353 |
586 |
763 |
1172 |
1758 |
|
Yeast6 |
1484 |
2.49 |
1188 |
30 |
1158 |
30 |
1.3% |
3 |
9 |
18 |
28 |
40 |
54 |
107 |
177 |
230 |
354 |
530 |
|
Yeast5 |
1484 |
2.96 |
1189 |
36 |
1153 |
36 |
1.56% |
3 |
7 |
15 |
23 |
33 |
45 |
89 |
147 |
191 |
293 |
440 |
|
Yeast4 |
1484 |
3.43 |
1188 |
41 |
1147 |
41 |
1.79% |
3 |
6 |
13 |
20 |
29 |
39 |
77 |
128 |
167 |
256 |
384 |
|
Yeast2vs8 |
482 |
4.15 |
387 |
17 |
370 |
17 |
2.3% |
3 |
5 |
10 |
16 |
22 |
30 |
60 |
100 |
129 |
199 |
298 |
|
Glass5 |
214 |
4.2 |
173 |
8 |
165 |
8 |
2.42% |
3 |
5 |
10 |
15 |
21 |
29 |
57 |
94 |
123 |
188 |
282 |
|
Abalone9vs18 |
731 |
5.65 |
586 |
34 |
552 |
34 |
3.08% |
3 |
4 |
8 |
12 |
17 |
23 |
45 |
74 |
96 |
148 |
221 |
|
Glass4 |
214 |
6.07 |
172 |
11 |
161 |
11 |
3.42% |
3 |
4 |
7 |
11 |
15 |
20 |
40 |
67 |
87 |
133 |
199 |
|
Ecoli4 |
336 |
6.74 |
270 |
19 |
251 |
19 |
3.78% |
3 |
3 |
6 |
10 |
14 |
18 |
36 |
60 |
78 |
120 |
180 |
|
Glass2 |
214 |
8.78 |
173 |
16 |
157 |
16 |
5.1% |
3 |
3 |
5 |
7 |
10 |
14 |
27 |
45 |
58 |
89 |
133 |
|
Vowel0 |
988 |
9.01 |
792 |
72 |
720 |
72 |
5% |
3 |
3 |
5 |
7 |
10 |
14 |
28 |
45 |
59 |
90 |
135 |
|
Page-blocks0 |
5472 |
10.23 |
4378 |
448 |
3930 |
448 |
5.7% |
3 |
3 |
4 |
7 |
9 |
12 |
24 |
40 |
52 |
79 |
118 |
|
Ecoli3 |
336 |
10.88 |
270 |
30 |
240 |
30 |
6.25% |
3 |
3 |
4 |
6 |
8 |
11 |
22 |
36 |
47 |
72 |
108 |
|
Yeast3 |
1484 |
10.98 |
1188 |
131 |
1057 |
131 |
6.2% |
3 |
3 |
4 |
6 |
8 |
11 |
22 |
36 |
47 |
72 |
108 |
|
Glass6 |
214 |
13.55 |
173 |
24 |
149 |
24 |
8.05% |
3 |
3 |
3 |
5 |
7 |
9 |
17 |
28 |
36 |
55 |
83 |
|
Segment0 |
2308 |
14.26 |
1848 |
264 |
1584 |
264 |
8.33% |
3 |
3 |
3 |
5 |
6 |
8 |
16 |
27 |
35 |
53 |
80 |
|
Ecoli2 |
336 |
15.48 |
270 |
42 |
228 |
42 |
9.21% |
3 |
3 |
3 |
4 |
6 |
8 |
15 |
24 |
32 |
48 |
72 |
|
New-thyroid1 |
215 |
16.28 |
173 |
29 |
144 |
29 |
10.07% |
3 |
3 |
3 |
4 |
5 |
7 |
14 |
22 |
29 |
44 |
66 |
|
New-thyroid2 |
215 |
16.89 |
173 |
30 |
143 |
30 |
10.49% |
3 |
3 |
3 |
4 |
5 |
7 |
13 |
21 |
28 |
42 |
63 |
|
Ecoli1 |
336 |
22.92 |
270 |
62 |
208 |
62 |
14.9% |
3 |
3 |
3 |
3 |
4 |
5 |
9 |
15 |
19 |
29 |
43 |
|
Vehicle0 |
846 |
23.64 |
677 |
160 |
517 |
160 |
15.47% |
3 |
3 |
3 |
3 |
4 |
5 |
9 |
14 |
18 |
28 |
42 |
|
Glass0123vs456 |
214 |
23.83 |
172 |
41 |
131 |
41 |
15.65% |
3 |
3 |
3 |
3 |
4 |
5 |
9 |
14 |
18 |
28 |
41 |
|
Haberman |
306 |
27.42 |
246 |
68 |
178 |
68 |
19.1% |
3 |
3 |
3 |
3 |
3 |
4 |
7 |
11 |
15 |
22 |
33 |
|
Vehicle1 |
846 |
28.37 |
678 |
193 |
485 |
193 |
19.9% |
3 |
3 |
3 |
3 |
3 |
4 |
7 |
11 |
14 |
21 |
32 |
|
Vehicle2 |
846 |
28.37 |
678 |
193 |
485 |
193 |
19.9% |
3 |
3 |
3 |
3 |
3 |
4 |
7 |
11 |
14 |
21 |
32 |
|
Vehicle3 |
846 |
28.37 |
678 |
193 |
485 |
193 |
19.9% |
3 |
3 |
3 |
3 |
3 |
4 |
7 |
11 |
14 |
21 |
32 |
|
Yeast1 |
1484 |
28.91 |
1188 |
344 |
844 |
344 |
20.38% |
3 |
3 |
3 |
3 |
3 |
4 |
7 |
11 |
14 |
21 |
31 |
|
Glass0 |
214 |
32.71 |
172 |
56 |
116 |
56 |
24.14% |
3 |
3 |
3 |
3 |
3 |
3 |
6 |
9 |
11 |
17 |
26 |
|
Iris0 |
150 |
33.33 |
121 |
40 |
81 |
40 |
24.69% |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
9 |
11 |
17 |
25 |
|
Pima |
768 |
34.84 |
616 |
215 |
401 |
215 |
26.81% |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
8 |
10 |
15 |
23 |
|
Ecoli0vs1 |
220 |
35 |
177 |
62 |
115 |
62 |
26.96% |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
8 |
10 |
15 |
22 |
|
Wisconsin |
683 |
35 |
548 |
192 |
356 |
192 |
26.97% |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
8 |
10 |
15 |
22 |
|
Glass1 |
214 |
35.51 |
172 |
61 |
111 |
61 |
27.48% |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
8 |
10 |
15 |
22 |
|
Mean |
919.94 |
17.61 |
737.09 |
96.61 |
640.48 |
96.61 |
12.02% |
3 |
4 |
7 |
10 |
13 |
18 |
35 |
58 |
75 |
115 |
172 |
|
StdDev |
1151.99 |
11.71 |
921.47 |
105.74 |
863.98 |
105.74 |
9% |
0 |
4.30 |
9.81 |
15.93 |
23 |
31.41 |
62.77 |
104.29 |
135.81 |
208.72 |
313.07 |
|
Median |
482 |
15.48 |
387 |
42 |
356 |
42 |
9.21% |
3 |
3 |
3 |
4 |
6 |
8 |
15 |
24 |
32 |
48 |
72 |
Table 5: sizeOfMinClass sized
subsample amounts for imbalanced data sets
|
|
Original |
Training sample |
Subsample |
|
Coverage maxSize |
|||||||||||||
|
Data set |
Size |
%Min |
Size |
Min.
Class Size |
Maj.
Class Size |
Size |
|
N_S |
||||||||||
|
|
|
|
|
|
N_S=3 |
10% |
20% |
30% |
40% |
50% |
75% |
90% |
95% |
99% |
99.9% |
|||
|
Abalone19 |
4174 |
0.77 |
3340 |
26 |
3314 |
52 |
0.78% |
3 |
14 |
29 |
46 |
65 |
89 |
177 |
293 |
381 |
585 |
878 |
|
Yeast6 |
1484 |
2.49 |
1188 |
30 |
1158 |
60 |
2.59% |
3 |
5 |
9 |
14 |
20 |
27 |
53 |
88 |
115 |
176 |
264 |
|
Yeast5 |
1484 |
2.96 |
1189 |
36 |
1153 |
72 |
3.12% |
3 |
4 |
8 |
12 |
17 |
22 |
44 |
73 |
95 |
146 |
218 |
|
Yeast4 |
1484 |
3.43 |
1188 |
41 |
1147 |
82 |
3.57% |
3 |
3 |
7 |
10 |
15 |
20 |
39 |
64 |
83 |
127 |
190 |
|
Yeast2vs8 |
482 |
4.15 |
387 |
17 |
370 |
34 |
4.59% |
3 |
3 |
5 |
8 |
11 |
15 |
30 |
49 |
64 |
98 |
147 |
|
Glass5 |
214 |
4.2 |
173 |
8 |
165 |
16 |
4.85% |
3 |
3 |
5 |
8 |
11 |
14 |
28 |
47 |
61 |
93 |
139 |
|
Abalone9vs18 |
731 |
5.65 |
586 |
34 |
552 |
68 |
6.16% |
3 |
3 |
4 |
6 |
9 |
11 |
22 |
37 |
48 |
73 |
109 |
|
Glass4 |
214 |
6.07 |
172 |
11 |
161 |
22 |
6.83% |
3 |
3 |
4 |
6 |
8 |
10 |
20 |
33 |
43 |
66 |
98 |
|
Ecoli4 |
336 |
6.74 |
270 |
19 |
251 |
38 |
7.57% |
3 |
3 |
3 |
5 |
7 |
9 |
18 |
30 |
39 |
59 |
88 |
|
Glass2 |
214 |
8.78 |
173 |
16 |
157 |
32 |
10.19% |
3 |
3 |
3 |
4 |
5 |
7 |
13 |
22 |
28 |
43 |
65 |
|
Vowel0 |
988 |
9.01 |
792 |
72 |
720 |
144 |
10% |
3 |
3 |
3 |
4 |
5 |
7 |
14 |
22 |
29 |
44 |
66 |
|
Page-blocks0 |
5472 |
10.23 |
4378 |
448 |
3930 |
896 |
11.4% |
3 |
3 |
3 |
3 |
5 |
6 |
12 |
20 |
25 |
39 |
58 |
|
Ecoli3 |
336 |
10.88 |
270 |
30 |
240 |
60 |
12.5% |
3 |
3 |
3 |
3 |
4 |
6 |
11 |
18 |
23 |
35 |
52 |
|
Yeast3 |
1484 |
10.98 |
1188 |
131 |
1057 |
262 |
12.39% |
3 |
3 |
3 |
3 |
4 |
6 |
11 |
18 |
23 |
35 |
53 |
|
Glass6 |
214 |
13.55 |
173 |
24 |
149 |
48 |
16.11% |
3 |
3 |
3 |
3 |
3 |
4 |
8 |
14 |
18 |
27 |
40 |
|
Segment0 |
2308 |
14.26 |
1848 |
264 |
1584 |
528 |
16.67% |
3 |
3 |
3 |
3 |
3 |
4 |
8 |
13 |
17 |
26 |
38 |
|
Ecoli2 |
336 |
15.48 |
270 |
42 |
228 |
84 |
18.42% |
3 |
3 |
3 |
3 |
3 |
4 |
7 |
12 |
15 |
23 |
34 |
|
New-thyroid1 |
215 |
16.28 |
173 |
29 |
144 |
58 |
20.14% |
3 |
3 |
3 |
3 |
3 |
4 |
7 |
11 |
14 |
21 |
31 |
|
New-thyroid2 |
215 |
16.89 |
173 |
30 |
143 |
60 |
20.98% |
3 |
3 |
3 |
3 |
3 |
3 |
6 |
10 |
13 |
20 |
30 |
|
Ecoli1 |
336 |
22.92 |
270 |
62 |
208 |
124 |
29.81% |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
7 |
9 |
14 |
20 |
|
Vehicle0 |
846 |
23.64 |
677 |
160 |
517 |
320 |
30.95% |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
7 |
9 |
13 |
19 |
|
Glass0123vs456 |
214 |
23.83 |
172 |
41 |
131 |
82 |
31.3% |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
7 |
8 |
13 |
19 |
|
Haberman |
306 |
27.42 |
246 |
68 |
178 |
136 |
38.2% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
7 |
10 |
15 |
|
Vehicle1 |
846 |
28.37 |
678 |
193 |
485 |
386 |
39.79% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
6 |
10 |
14 |
|
Vehicle2 |
846 |
28.37 |
678 |
193 |
485 |
386 |
39.79% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
6 |
10 |
14 |
|
Vehicle3 |
846 |
28.37 |
678 |
193 |
485 |
386 |
39.79% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
6 |
10 |
14 |
|
Yeast1 |
1484 |
28.91 |
1188 |
344 |
844 |
688 |
40.76% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
5 |
6 |
9 |
14 |
|
Glass0 |
214 |
32.71 |
172 |
56 |
116 |
112 |
48.28% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
5 |
7 |
11 |
|
Iris0 |
150 |
33.33 |
121 |
40 |
81 |
80 |
49.38% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
5 |
7 |
11 |
|
Pima |
768 |
34.84 |
616 |
215 |
401 |
430 |
53.62% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
6 |
9 |
|
Ecoli0vs1 |
220 |
35 |
177 |
62 |
115 |
124 |
53.91% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
6 |
9 |
|
Wisconsin |
683 |
35 |
548 |
192 |
356 |
384 |
53.93% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
6 |
9 |
|
Glass1 |
214 |
35.51 |
172 |
61 |
111 |
122 |
54.95% |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
6 |
9 |
|
Mean |
919.94 |
17.61 |
737.09 |
96.61 |
640.48 |
193.21 |
24.04% |
3 |
3 |
4 |
6 |
7 |
9 |
17 |
28 |
37 |
56 |
84 |
|
StdDev |
1151.99 |
11.71 |
921.47 |
105.74 |
863.98 |
211.47 |
18% |
0 |
1.94 |
4.66 |
7.76 |
11.27 |
15.56 |
31.47 |
52.23 |
68 |
104.38 |
156.68 |
|
Median |
482 |
15.48 |
387 |
42 |
356 |
84 |
18.42% |
3 |
3 |
3 |
3 |
3 |
4 |
7 |
12 |
15 |
23 |
34 |
Table 6: maxSize sized subsample amounts for
imbalanced data sets
3 Results
of the Wilcoxon Signed Ranks test regarding comparisons between subsample sizes
The tables in this section show the results of the Wilcoxon tests
applied to look for statistically significant differences between CTC using
different subsample sizes for the same coverage. Each table represents the
results of one of the classification contexts. Table 7 represents standard classification, Table 8 represents imbalanced classification and Table
9 represents the classification of imbalanced
data sets preprocessed with SMOTE. In these tables refers to the ranking of sizeOfMinClass
subsamples while refers to maxSize subsamples, with the
higher rank stressed in bold. Each of the tables is followed a by a figure
graphically showing the average performance of CTC for that context.
|
Measure |
Coverage |
p-value |
Hypothesis(α = 0.05) |
||
|
Kappa |
N_S=3 |
212 |
253 |
0.673 |
Not rejected |
|
Kappa |
10% |
199 |
266 |
0.491 |
Not rejected |
|
Kappa |
20% |
190 |
275 |
0.982 |
Not rejected |
|
Kappa |
30% |
193 |
272 |
0.417 |
Not rejected |
|
Kappa |
40% |
216 |
249 |
0.734 |
Not rejected |
|
Kappa |
50% |
174 |
291 |
0.229 |
Not rejected |
|
Kappa |
75% |
216 |
249 |
0.734 |
Not rejected |
|
Kappa |
90% |
220 |
245 |
0.797 |
Not rejected |
|
Kappa |
95% |
273 |
192 |
0.405 |
Not rejected |
|
Kappa |
99% |
208 |
257 |
0.614 |
Not rejected |
|
Kappa |
99.9% |
223 |
242 |
0.845 |
Not rejected |
|
Accuracy |
N_S=3 |
221 |
244 |
0.813 |
Not rejected |
|
Accuracy |
10% |
206 |
259 |
0.586 |
Not rejected |
|
Accuracy |
20% |
206 |
259 |
0.586 |
Not rejected |
|
Accuracy |
30% |
217 |
248 |
0.750 |
Not rejected |
|
Accuracy |
40% |
215 |
250 |
0.719 |
Not rejected |
|
Accuracy |
50% |
206 |
259 |
0.586 |
Not rejected |
|
Accuracy |
75% |
223 |
242 |
0.845 |
Not rejected |
|
Accuracy |
90% |
233 |
232 |
0.992 |
Not rejected |
|
Accuracy |
95% |
277 |
188 |
0.360 |
Not rejected |
|
Accuracy |
99% |
215 |
250 |
0.719 |
Not rejected |
|
Accuracy |
99.9% |
232 |
233 |
0.992 |
Not rejected |
Table 7: Wilcoxon test comparing
differences for kappa and accuracy for different subsample sizes over standard
data sets.
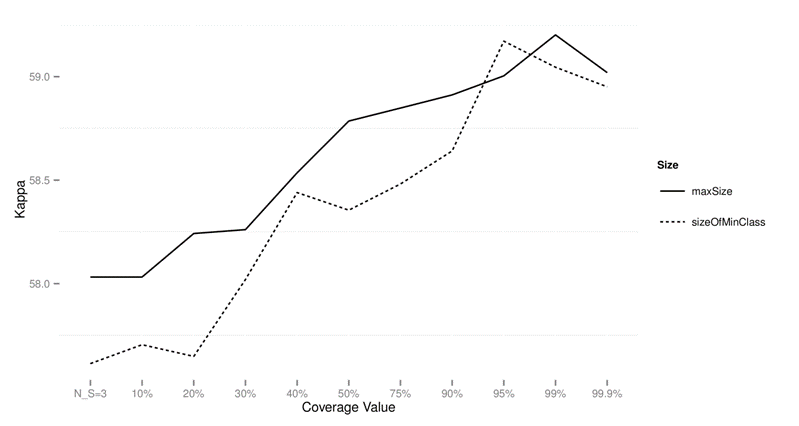
Figure 1: Performance of CTC with
different subsample sizes for different values of data sets on standard data
sets using kappa as the performance measure.
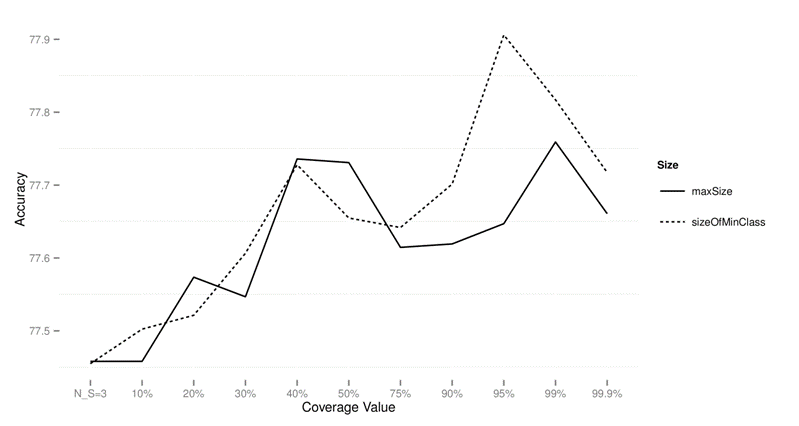
Figure 2: Performance of CTC with
different subsample sizes for different values of data sets on standard data
sets using accuracy as the performance measure.
|
Measure |
Coverage |
p-value |
Hypothesis(α = 0.05) |
||
|
GM |
N_S=3 |
121 |
439 |
0.004 |
Rejected in favor of maxSize |
|
GM |
10% |
134 |
426 |
0.009 |
Rejected in favor of maxSize |
|
GM |
20% |
146 |
414 |
0.016 |
Rejected in favor of maxSize |
|
GM |
30% |
114 |
396 |
0.037 |
Rejected in favor of maxSize |
|
GM |
40% |
165 |
395 |
0.039 |
Rejected in favor of maxSize |
|
GM |
50% |
174 |
386 |
0.057 |
Not rejected |
|
GM |
75% |
155 |
405 |
0.025 |
Rejected in favor of maxSize |
|
GM |
90% |
171 |
389 |
0.050 |
Rejected in favor of maxSize |
|
GM |
95% |
169 |
391 |
0.046 |
Rejected in favor of maxSize |
|
GM |
99% |
203 |
357 |
0.166 |
Not rejected |
|
GM |
99.9% |
148 |
412 |
0.018 |
Rejected in favor of maxSize |
Table 8: Wilcoxon test comparing differences
for GM for different subsample sizes over imbalanced data sets.
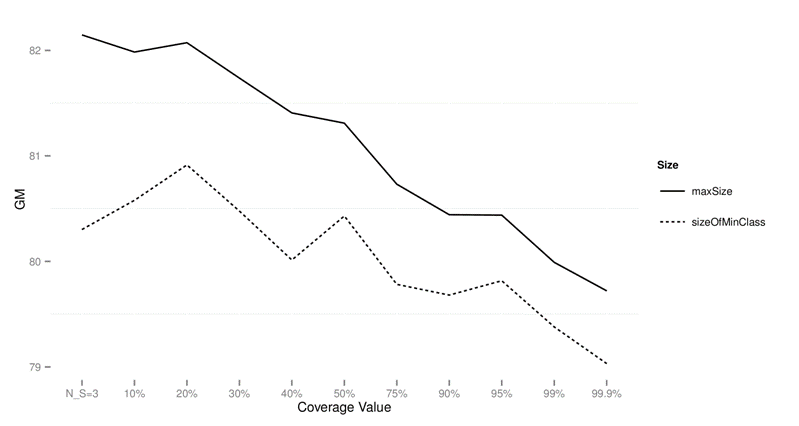
Figure 3: Performance of CTC with
different subsample sizes for different values of data sets on imbalanced data
sets using GM as the performance measure.
|
Measure |
Coverage |
p-value |
Hypothesis(α = 0.05) |
||
|
F1-Score |
N_S=3 |
97 |
463 |
0,001 |
Rejected in favor of maxSize |
|
F1-Score |
10% |
101 |
459 |
0,001 |
Rejected in favor of maxSize |
|
F1-Score |
20% |
120 |
440 |
0,004 |
Rejected in favor of maxSize |
|
F1-Score |
30% |
152 |
408 |
0,022 |
Rejected in favor of maxSize |
|
F1-Score |
40% |
161 |
399 |
0,033 |
Rejected in favor of maxSize |
|
F1-Score |
50% |
220 |
340 |
0,28 |
Not rejected |
|
F1-Score |
75% |
189 |
371 |
0,102 |
Not rejected |
|
F1-Score |
90% |
205 |
355 |
0,177 |
Not rejected |
|
F1-Score |
95% |
243 |
317 |
0,503 |
Not rejected |
|
F1-Score |
99% |
230 |
330 |
0,367 |
Not rejected |
|
F1-Score |
99.9% |
160 |
400 |
0,031 |
Rejected in favor of maxSize |
Table 10:
Wilcoxon test comparing differences for F1-Score for different subsample sizes
over imbalanced data sets.
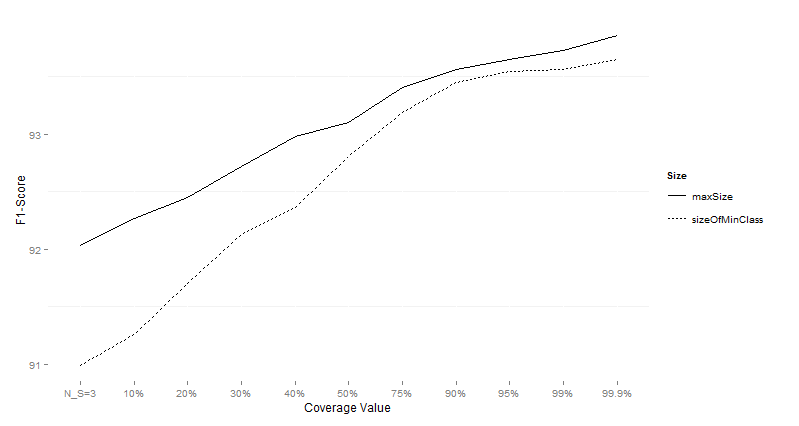
Figure 4:
Performance of CTC with different subsample sizes for different values of data
sets on imbalanced data sets using the F1-Score as the performance measure.
|
Measure |
Coverage |
p-value |
Hypothesis(α = 0.05) |
||
|
MCC |
N_S=3 |
38 |
522 |
0,00001 |
Rejected in favor of maxSize |
|
MCC |
10% |
43 |
517 |
0,00002 |
Rejected in favor of maxSize |
|
MCC |
20% |
52 |
508 |
0,00004 |
Rejected in favor of maxSize |
|
MCC |
30% |
106 |
454 |
0,002 |
Rejected in favor of maxSize |
|
MCC |
40% |
85 |
475 |
0,001 |
Rejected in favor of maxSize |
|
MCC |
50% |
144 |
416 |
0,015 |
Rejected in favor of maxSize |
|
MCC |
75% |
112 |
448 |
0,003 |
Rejected in favor of maxSize |
|
MCC |
90% |
147 |
413 |
0,017 |
Rejected in favor of maxSize |
|
MCC |
95% |
225 |
335 |
0,321 |
Not rejected |
|
MCC |
99% |
177 |
383 |
0,064 |
Not
rejected |
|
MCC |
99.9% |
121 |
439 |
0,004 |
Rejected in favor of maxSize |
Table 10:
Wilcoxon test comparing differences for MCC for different subsample sizes over
imbalanced data sets.
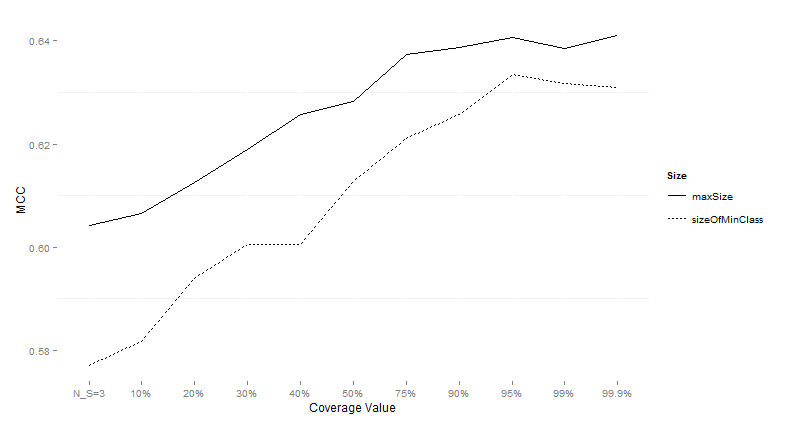
Figure 5:
Performance of CTC with different subsample sizes for different values of data
sets on imbalanced data sets using MCC as the performance measure.
|
Measure |
Coverage |
p-value |
Hypothesis(α = 0.05) |
||
|
GM |
N_S=3 |
197 |
364 |
0,136 |
Not rejected |
|
GM |
10% |
236 |
325 |
0,427 |
Not rejected |
|
GM |
20% |
206 |
355 |
0,183 |
Not rejected |
|
GM |
30% |
181 |
380 |
0,075 |
Not rejected |
|
GM |
40% |
228 |
333 |
0,348 |
Not rejected |
|
GM |
50% |
222 |
339 |
0,296 |
Not rejected |
|
GM |
75% |
272 |
289 |
0,879 |
Not rejected |
|
GM |
90% |
220 |
341 |
0,280 |
Not rejected |
|
GM |
95% |
310 |
251 |
0,598 |
Not rejected |
|
GM |
99% |
264 |
297 |
0,768 |
Not rejected |
|
GM |
99.9% |
361 |
200 |
0,150 |
Not rejected |
Table 9: Wilcoxon test comparing
differences for GM for different subsample sizes over imbalanced data sets
preprocessed with SMOTE.
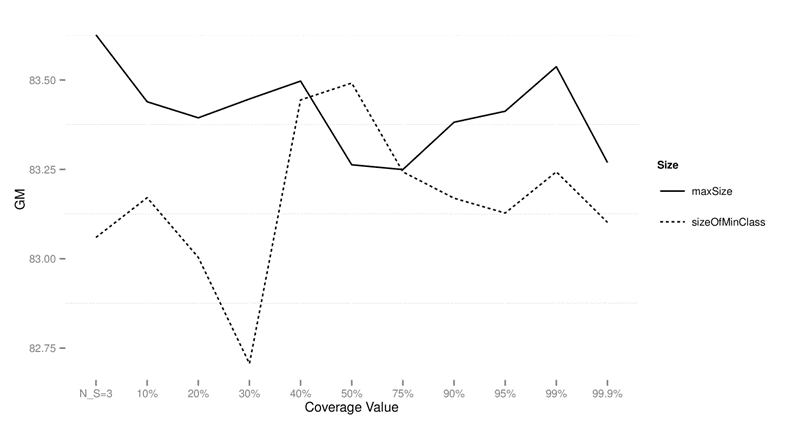
Figure 6:
Performance of CTC with different subsample sizes for different values of data sets
on imbalanced data sets preprocessed with SMOTE using GM as the performance
measure.
|
Measure |
Coverage |
p-value |
Hypothesis(α = 0.05) |
||
|
F1-Score |
N_S=3 |
49 |
512 |
0,0004 |
Rejected in favor of maxSize |
|
F1-Score |
10% |
86 |
485 |
0,0003 |
Rejected in favor of maxSize |
|
F1-Score |
20% |
46 |
515 |
0,0003 |
Rejected in favor of maxSize |
|
F1-Score |
30% |
33 |
528 |
0,0001 |
Rejected in favor of maxSize |
|
F1-Score |
40% |
58 |
503 |
0,001 |
Rejected in favor of maxSize |
|
F1-Score |
50% |
49 |
512 |
0,0004 |
Rejected in favor of maxSize |
|
F1-Score |
75% |
94 |
467 |
0,001 |
Rejected in favor of maxSize |
|
F1-Score |
90% |
78 |
483 |
0,0003 |
Rejected in favor of maxSize |
|
F1-Score |
95% |
50 |
511 |
0,0004 |
Rejected in favor of maxSize |
|
F1-Score |
99% |
92 |
469 |
0,001 |
Rejected in favor of maxSize |
|
F1-Score |
99.9% |
102 |
459 |
0,001 |
Rejected in favor of maxSize |
Table 10:
Wilcoxon test comparing differences for F1-Score for different subsample sizes
over imbalanced data sets preprocessed with SMOTE.
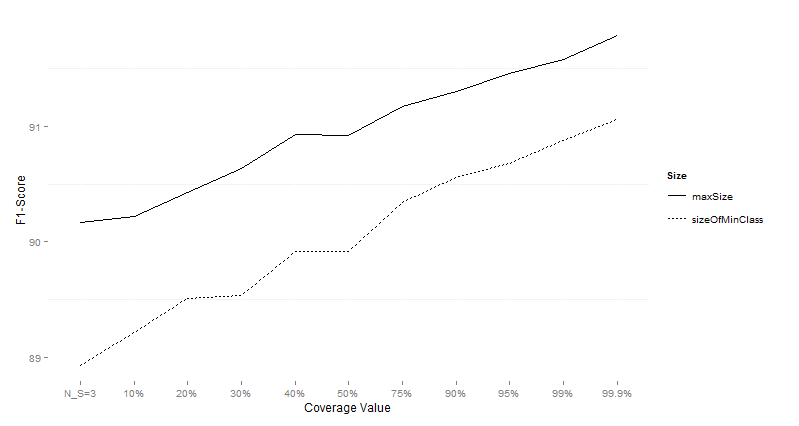
Figure 7:
Performance of CTC with different subsample sizes for different values of data sets
on imbalanced data sets preprocessed with SMOTE using the F1-Score as the
performance measure.
|
Measure |
Coverage |
p-value |
Hypothesis(α = 0.05) |
||
|
MCC |
N_S=3 |
140 |
421 |
0,012 |
Rejected in favor of maxSize |
|
MCC |
10% |
167 |
394 |
0,043 |
Rejected in favor of maxSize |
|
MCC |
20% |
132 |
429 |
0,008 |
Rejected in favor of maxSize |
|
MCC |
30% |
100 |
461 |
0,001 |
Rejected in favor of maxSize |
|
MCC |
40% |
148 |
413 |
0,018 |
Rejected in favor of maxSize |
|
MCC |
50% |
147 |
414 |
0,017 |
Rejected in favor of maxSize |
|
MCC |
75% |
185 |
376 |
0,088 |
Not rejected |
|
MCC |
90% |
140 |
421 |
0,012 |
Rejected in favor of maxSize |
|
MCC |
95% |
141 |
420 |
0,013 |
Rejected in favor of maxSize |
|
MCC |
99% |
158 |
403 |
0,029 |
Rejected in favor of maxSize |
|
MCC |
99.9% |
171 |
390 |
0,051 |
Not rejected |
Table 11:
Wilcoxon test comparing differences for MCC for different subsample sizes over imbalanced
data sets preprocessed with SMOTE.
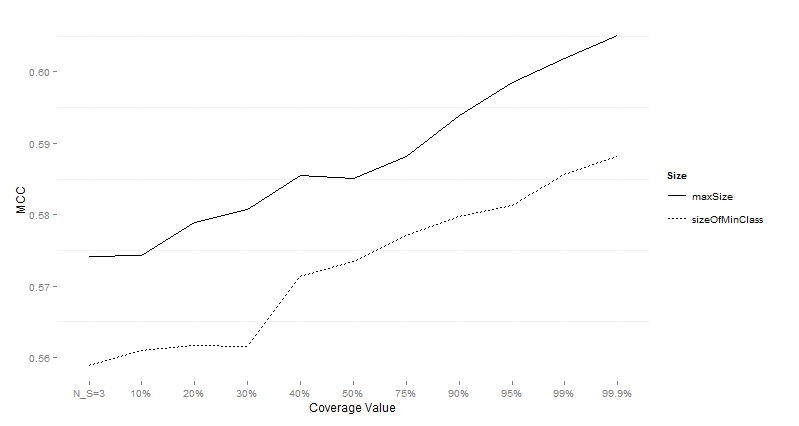
Figure 8:
Performance of CTC with different subsample sizes for different values of data
sets on imbalanced data sets preprocessed with SMOTE using MCC as the
performance measure.
4 Execution times for CTC
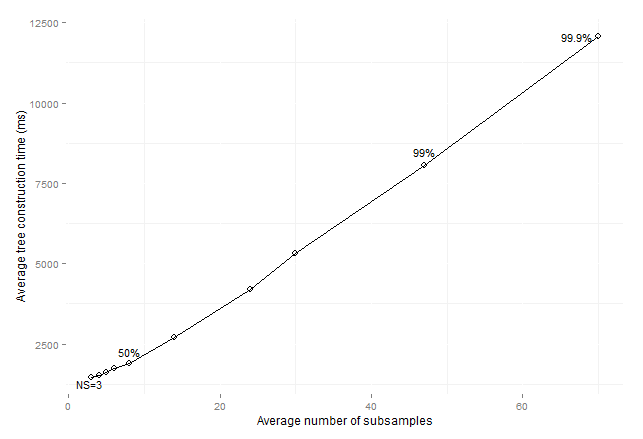
Figure 9: Average consolidated
tree construction time by average number of subsamples for standard data sets (sizeOfMinClass subsamples).
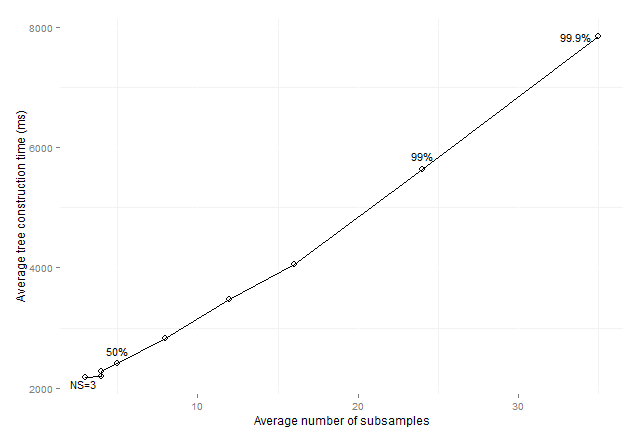
Figure 10: Average
consolidated tree construction time by average number of subsamples for
standard data sets (maxSize
subsamples).
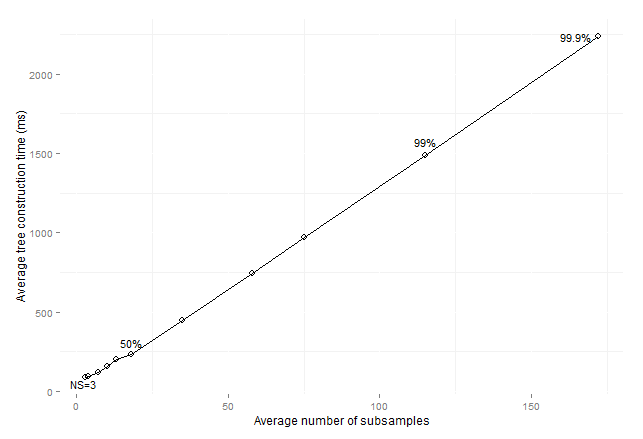
Figure 11: Average
consolidated tree construction time by average number of subsamples for
imbalanced data sets (sizeOfMinClass
subsamples).
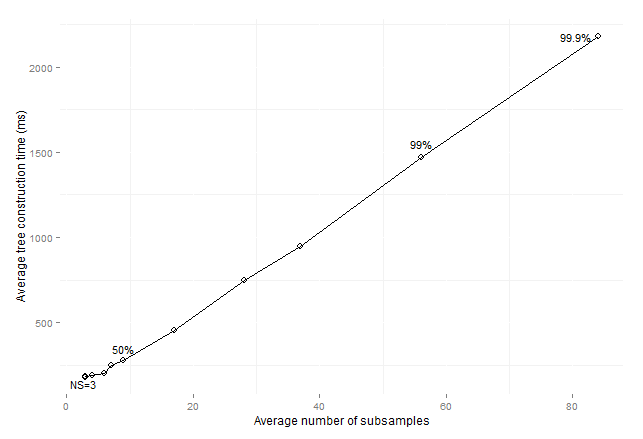
Figure 12: Average
consolidated tree construction time by average number of subsamples for
imbalanced data sets (maxSize
subsamples).
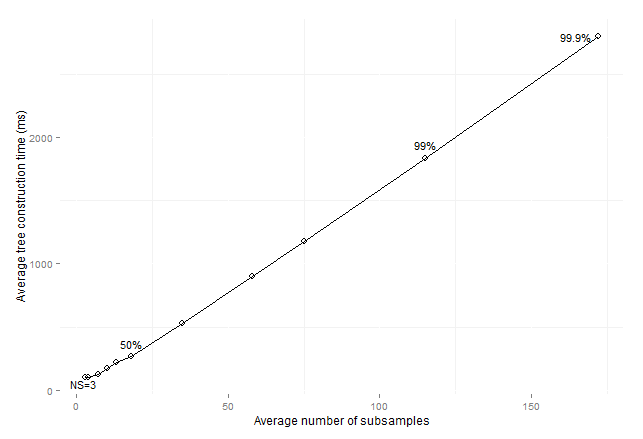
Figure 13: Average consolidated
tree construction time by average number of subsamples for imbalanced data sets
preprocessed with SMOTE (sizeOfMinClass
subsamples).
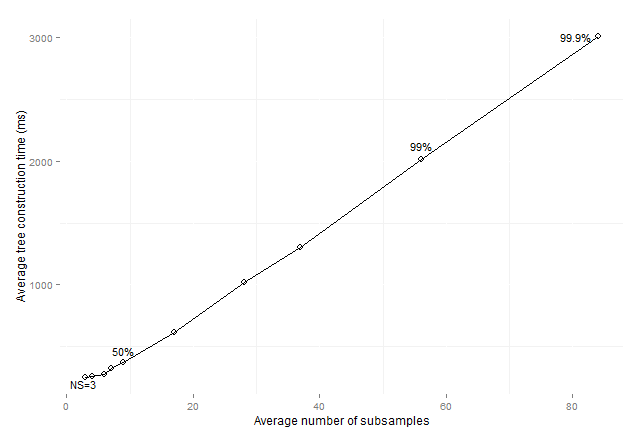
Figure 14: Average consolidated
tree construction time by average number of subsamples for imbalanced data sets
preprocessed with SMOTE (maxSize
subsamples).
5
Average Results achieved by CTC
For the
sake of replicability we publish the average results obtained by CTC for both
subsample sizes on all three classification contexts.
·
Standard
classification. SizeOfMinClass subsample size. Kappa performance measure. ![]() and
and ![]()
·
Standard
classification. maxSize subsample size. Kappa performance measure. ![]() and
and ![]()
·
Standard
classification. SizeOfMinClass subsample size. Accuracy performance measure. ![]() and
and ![]()
·
Standard
classification. maxSize subsample size. Accuracy performance measure. ![]() and
and ![]()
·
Imbalanced
classification. SizeOfMinClass subsample size. GM performance measure. ![]() and
and ![]()
·
Imbalanced
classification. maxSize subsample size. GM performance measure. ![]() and
and ![]()
·
Imbalanced
classification preprocessed with SMOTE. SizeOfMinClass subsample size. GM
performance measure. ![]() and
and ![]()
·
Imbalanced
classification preprocessed with SMOTE. maxSize subsample size. GM performance
measure. ![]() and
and ![]()
6 Comparison between CTC and methods to tackle
class imbalance
·
Imbalanced
classification. CTC and SMOTE+CTC vs 8 methods. AUC performance measure. ![]() and
and ![]()