The datasets included below are the adapted versions of the original spectra data available on each authors' websites. Notice that the downloadable files are Matlab workspaces files, including three variables: 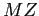 , the values of the
, the values of the 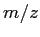 axis;
axis; 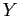 , the spectra intensity values for each
, the spectra intensity values for each 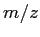 position;
position; 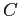 the phenotypic distribution of the spectra included in
the phenotypic distribution of the spectra included in 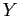 .
.
- Ovarian cancer profiling (OVA) [Petricoin et al. (2002)]. Being one of the pioneering works on MS data profiling from serum samples, the work by Petricoin et al. (2002) is now one of the most analysed benchmark MS datasets. The aim is to separate serum samples of a female population with ovarian cancer from control samples of unaffected women using a small set of proteomic markers. The available data contain 200 SELDI spectra of 121 cancer samples and 79 controls. The
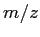 values range from 700.116 to 12,000 with a total of 45,200 values per spectrum.
values range from 700.116 to 12,000 with a total of 45,200 values per spectrum.
- Detection of drug-induced toxicity (TOX) [Petricoin et al. (2004)]. In this work, rat models are analysed using a serum proteomic pattern diagnostic device based on a SELDI-TOF spectrometer. The study intends to find biomarkers able to distinguish between anthracycline- and anthracenedione-induced cardiotoxicity and control samples. The separation of the training and test sets in the original work is confusing. Consequently, we just picked the samples diagnosed as definite positive or definite negative. Our TOX dataset then is composed of 62 samples of two phenotypes with 28 and 34 samples each. As in the previous dataset, a total of 45,200
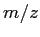 values are configured, ranging from 799.115 to 12,000.
values are configured, ranging from 799.115 to 12,000.
- Hepatocellular carcinoma (HCC) [Ressom et al. (2006)].This study sets out to help discover early markers for hepatocellular carcinomas triggered by viral infections. The samples were obtained from the Kasr El-Aini Hospital (Cairo, Egypt), where this carcinoma is a primary health problem. After removing proteins greater than 50 kDA (including albumin), the spectra are generated by a MALDI-TOF instrument. The dataset includes 36,802
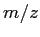 final readings for 150 samples, 78 affected and 72 non-affected controls.
final readings for 150 samples, 78 affected and 72 non-affected controls.
- Detection of glycan biomarkers (DGB) [Ressom et al. (2008)]. The original work proposes a method for systematically selecting glycan structures able to distinguish subjects from pre-labeled groups. Over a set of three different phenotypes, the glycans are released from their associated proteins through an enzymatic treatment and later methylated to avoid solubility. The available data comprises a total of 128 MALDI-TOF spectra: 78 from healthy controls, 25 from hepatocellular carcinoma and another 25 from chronic liver disease samples. The
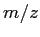 values are in the 1,499.8 to 5,518.3 interval, with a total of 16,075 points for each sample.
values are in the 1,499.8 to 5,518.3 interval, with a total of 16,075 points for each sample.